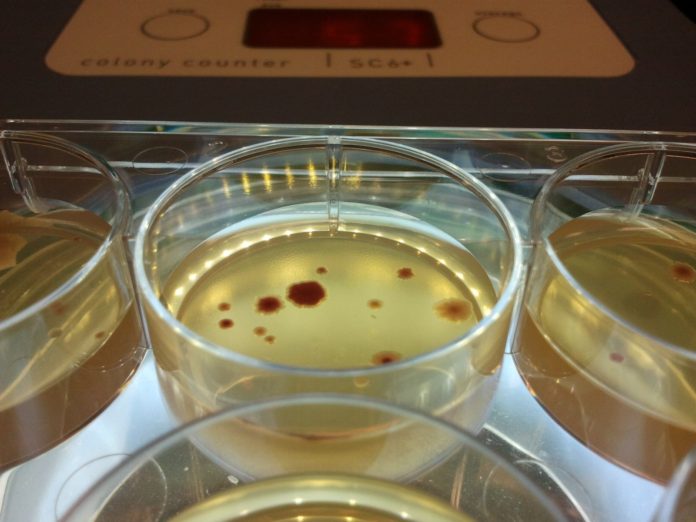
Lonely microbes are those that live at low population densities, and this month a study led by a team at The University of Manchester has demonstrated how they’re more likely to mutate causing antibiotic resistance levels to rise. The study has members of the team analyzing around 70 years worth of data and around 500 different mutation measurements. In doing this, they found that individual microbes, such as bacteria, that live in denser populations, are far less likely to mutate than those in sparser groups.
Becoming antibiotic resistant is just one outcome caused by mutations in bacteria, but it’s one of the worst. The researchers are hopeful this study will enable scientists to gain a deeper understanding of antibiotic resistance and one day combat the rise of these dreaded ‘superbugs’. This latest study is a collaboration with the Universities of Keele and Middlesex and adds to previous research involving the population density of microbes and how it affected the mutation rate of E. coli. The new research is similar but analyzes mutation rates from all different species of microbes and viruses, not just E.coli.
Throughout the study, nearly 2 trillion microbial cells were used and a result similar to that of the previous study was achieved. “Spontaneous mutations fuel evolution, but when that termination leads to something more serious, such as resistance to antibiotics, it becomes an issue. That is why the particular mutations we looked at in the lab are those related to antibiotic resistance,” said Dr. Chris Knight from the University’s Faculty of Science and Engineering and senior author of the study.”
According to statistics released by the World Health Organization (WHO), as many as 10 million people per year will die by 2050 if this level of antibiotic resistance continues. Experts at the organization also say that this is threatening our ability to be able to prevent and treat certain diseases, parasites, viruses, infections, etc. And, as such, would be a serious threat to global public health.
DAMP stands for ‘density-associated mutation-rate plasticity’ and is a way to measure how these mutations vary with certain environments. Results from the study showed that DAMP played a big part in reducing those mutations responsible for causing antibiotic resistance. Lead author of the study, Dr. Krasovec from the Faculty of Biology, Medicine, and Health, explains, “In our analyses, DAMP gives bacteria a lower chance of becoming antibiotic resistance at higher population densities. We anticipate that DAMP affects the course of evolution more generally and understanding its causes and effects will help understand and control evolution.”
More News to Read
- Meet the Engineer Who 3D Prints Revolutionary Human-Robotic Arms
- Meteor Shower From Dead Comet Sighted Again
- Will Tesla 3 Pre-orders See the Light of Day and Will This Car Create…
- Future NASA Study Will Focus on the Ocean Worlds of the Milky Way
- Quantum Computer Programming: What You Need to Learn to Get Started?